1. INTRODUCTION
Termites are pests that cause a great damage to wood by consuming cellulose as a main food. Generally, termites are known to inhabit subtropical and tropical regions. Two species of lower termites occur in Korea (Wonhoon et al., 2015), and they can be observed at Bukhan Mountain in Seoul, Korea (Cho et al., 2010). During the winter in Korea, the temperature can drop below zero degrees Celsius, so even if termites can avoid freezing in their shelters, they must be physiologically adapted to these low temperatures. Lower termites rely on symbiotic microbes in their gut to assist their digestion (Brune and Ohkuma, 2011). Therefore, some of these symbiotic microorganisms in the intestine need to support the metabolic activity of termites at low temperatures. In a previous study, we found that one of the bacterial species that forms a symbiotic relationship with the termite species Reticulitermes speratus KMT001, known as Elizabethkingia sp. BM10, produces β-glucosidase at 10°C (Lee et al., 2018). Elizabethkingia sp. BM10 has since been re-named Elizabethkingia miricola BM10. Because of the unique characteristics that help termites by producing cellulases at low temperatures, further physiological studies of the strains were necessary. In the study we report here, we investigated the physiological characteristics of E. miricola BM10.
The genus Elizabethkingia is found in a variety of places including people, plants, insects, and water. It is well known to pathogenic bacteria in human. The genus Elizabethkingia currently consists of three species (Bernardet et al., 2006; Ceyhan and Celik, 2011). Two species, E. meningoseptica and E. miricola, were first classified when the genus Elizabethkingia was reclassified from the genus Chryseobacterium, while the third species, E. anophelis, was classified following its isolation from the midgut of the mosquito Anopheles gambiae (Kämpfer et al., 2011; Kim et al., 2005). E. meningoseptica was first reported more than five decades ago, whereas E. miricola was isolated and classified from condensation water of the space station Mir, in 2003 (King, 1959; Li et al., 2003). An additional new species, E. endophytica, was suggested based on DNA–DNA hybridization results and some biochemical properties, but a later, whole-genome sequencing comparison suggested that the new species was a subjective synonym of E. anophelis (Doijad et al., 2016; Kämpfer et al., 2015). Therefore, the controversial species E. endophytica was eliminated from comparisons with the new strain, E. miricola BM10. In this study, genetic analysis including genome, fat content analysis, and various physiological metabolism analysis were performed and compared with three type strains in the genus Elizabethkingia to determine the taxonomic location of E. miricola BM10.
2. MATERIALS and METHODS
E. miricola BM10 was isolated from the hindgut of the termite species Reticulitermes speratus KMT001 (Cho et al., 2010). This bacterial strain was deposited at the Korean Collection for Type Culture, under the number KCTC 18449P; at the Korean Culture Collection of Microorganisms, under the number KCCM 76545; and at the National Institute of Technology and Evaluation in Japan, under the number NBRC 112444. E. miricola BM10 was cultured in 1% tryptone, 0.5% yeast extract, and 1% sodium carboxymethyl cellulose (TYE-CMC medium) at 26°C for 2 days. For plate cultures, 1.5% agar was added to TYE-CMC medium.
The testing of E. miricola BM10 bacterial cell physiology was performed by the Korean Culture Center of Microorganisms (Seoul, Korea), using the API® 20NE kit (bioMérieux, Inc., Durham, NC, USA). Cell growth, as based on doubling-time, was measured in tryptic soy broth (TSB, Difco™, Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
For fatty acid composition analysis, the four Elizabethkingia strains were grown on tryptic soy agar (TSA, Difco™, Becton, Dickinson and Company) at 28°C for 2 days. The fatty acid composition of the four strains was analyzed, according to the Miller method, by the Korean Culture Center of Microorganisms (Miller, 1982). Fatty acid composition was analyzed using the Agilent 6890 Gas Chromatograph (Agilent Technologies, Santa Clara, CA, USA) and HP-1 capillary column (Agilent Technologies). The results were analyzed Sherlock™ Microbial ID System software version 6.2 (MIDI, Inc., Newark, DE, USA). Peaks were identified by retention time, and the relative peak area was calculated by comparison with standard calibration solutions.
For the chromosomal DNA analysis, E. miricola BM10 was cultivated on TYE-CMC agar plates at 26°C for 2 days. The colonies on the plates were sampled for 16S rRNA sequence analysis and whole-genome sequencing.
For the initial identification of E. miricola BM10, the 16S rRNA gene sequence was analyzed using two primers: 27F (5′-AGAGTTTGATCNTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Kämpfer et al., 2011). The complete genome sequence of E. miricola BM10 was analyzed and deposited in the GenBank of the National Center for Biotechnology Information (NCBI) under the accession number CP011059 (Lee et al., 2015).The average nucleotide identity (ANI) of the whole genome sequence of E. miricola BM10 was calculated using the ANI calculator (http://enve-omics.ce.gatech.edu/ani/) with 70% minimum identity in alignment options by pairwise comparison with the reference genome sequences of three other type strains: E. anophelis R26 (GenBank assembly accession: GCA_000331815.1), E. meningoseptica ATCC 13253 (GenBank assembly accession: GCA_ 000367325.1), and E. miricola GTC 862 (GenBank assembly accession: GCA_000769445.1) (Rodriguez-R and Konstantinidis, 2016). Phylogenetic analysis was performed using MEGA 4 software (http://www.megasoftware.net/mega4/mega.html) using three methods: neighbor-joining method, maximum-parsimony method, and unweighted pair-group method with arithmetic mean with the bootstrap phylogeny test with 1000 replications. The 16S rRNA gene sequences were trimmed and used for the phylogenetic analysis.
3. RESULTS and DISCUSSION
The termite species R. speratus KMT001 occurs on Bukhan Mountain in Seoul, Korea (Cho et al., 2010). Symbiotic bacteria from the hindgut of R. speratus KMT001 were isolated and E. miricola BM10 (named Elizabethkingia sp. BM10 by Cho et al. (2010)) was one of 16 bacterial species isolated (Cho et al., 2010). The bacteria were Gram-negative, non-motile, and non-spore-forming rods (0.5 × 1 μm). Good growth was observed on TSA, TYE-CMC, and Luria-Bertani agar at 26°C, while very slow growth was observed at 4°C. The organism’s doubling-time in the exponential growth phase was 1.12 h at 26°C in TSB medium. Colonies were white-yellow, circular, and semitranslucent, and had entirely shiny edges.
E. miricola BM10 could not reduce nitrates to nitrites. It produced indole from tryptophan. Its glucose acidification properties were negative. The bacterium could produce β-glucosidase (esculin hydrolysis), protease (gelatin hydrolysis), β-galactosidase, and cytochrome oxidase, but did not produce arginine dihydrolase and urease. It could assimilate glucose, mannose, N-acetyl-glucosamine, and citrate, but not arabinose, mannitol, maltose, gluconate, caprate, adipate, malate, or phenyl acetate. The biochemical test results are summarized and any differences among the four species are shown in Table 1.
The fatty acid content of E. miricola BM10 was compared with that of the three type-strains in the genus Elizabethkingia: E. anophelis R26, E. meningoseptica ATCC 13253, and E. miricola GTC 862 (Table 2). The major fatty acids found in E. miricola BM10 were iso- C15:0, iso-C17:0 3-OH, and summed feature 3 (iso-C16:1 ω7c/C16:1ω6c). The iso-C17:0 3-OH content in E. miricola BM10 was higher compared with that of the other three Elizabethkingia strains, but the iso-C16:0 3-OH and summed feature 3 content in E. miricola BM10 was lower compared with that of the other strains.
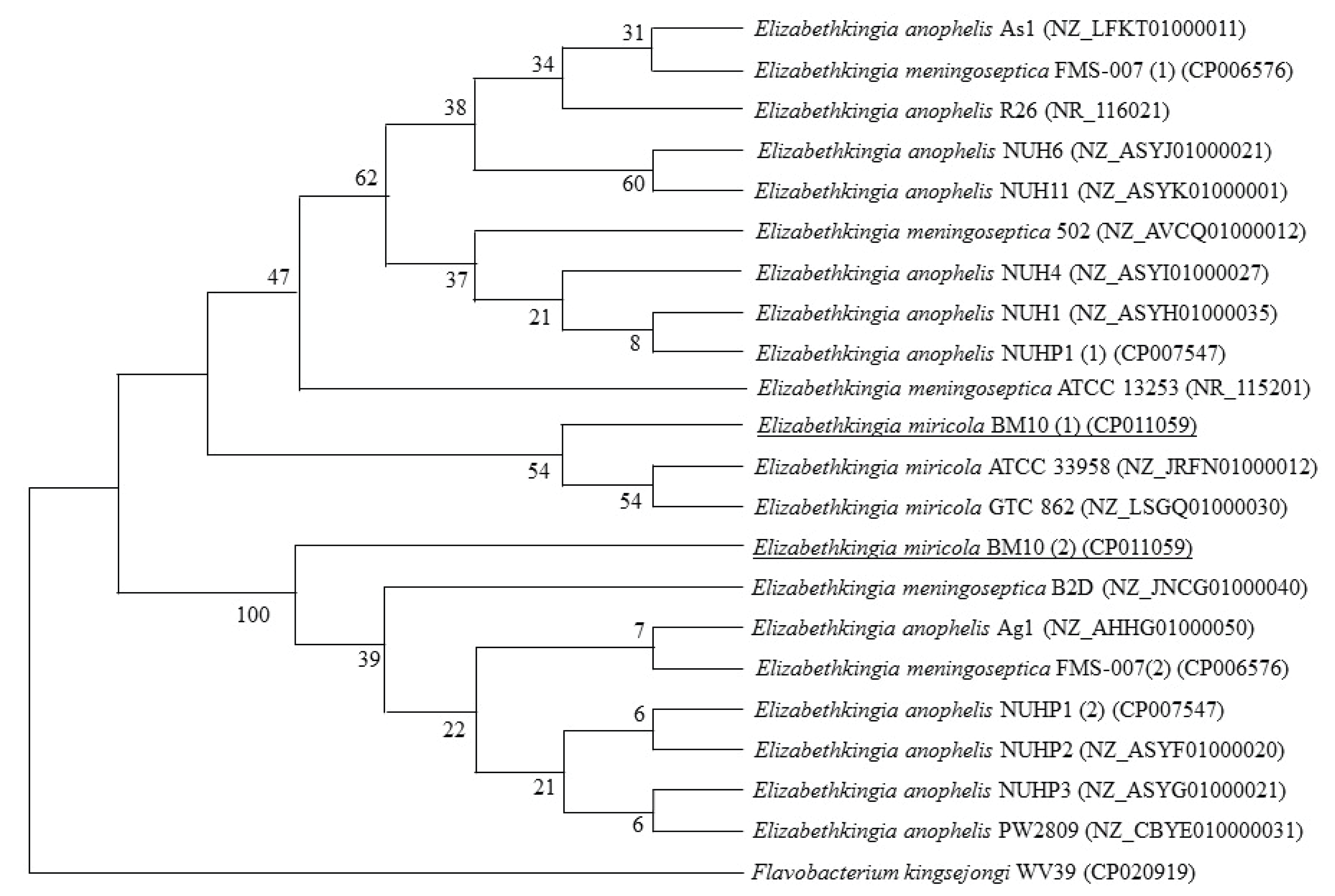
E. miricola BM10 had two 16S rRNA sequences, and their homology with the 16S rRNA sequences of type strains of the Elizabethkingia genus in the NCBI database was analyzed. The homology of the two 16S rRNA sequences of E. miricola BM10 were 98.3% and 98.7% with E. anophelis R26, 98.1% and 98.6% with E. meningoseptica ATCC 13253, and 99.4% and 99.4% with E. miricola GTC 862.
In the phylogenetic tree using the neighbor-joining method (Fig. 1), the first 16S rRNA sequences of E. miricola BM10 were grouped with two E. miricola strains, including E. miricola GTC 862, but were distinctly separate from the two strains of E. miricola. The second set of 16S rRNA sequences of E. miricola BM10 was separate from the rest of the sequences in its subgroup. An additional two phylogenetic analyses with the maximum-parsimony method and unweighted pair group method with arithmetic mean showed similar results (data not shown).
The whole genome of E. miricola BM10 was sequenced (Lee et al., 2015) and compared with three type-strains of the genus Elizabethkingia: E. anophelis R26, E. meningoseptica ATCC 13253, and E. miricola GTC 862 in Table 3 (Assembly ID in the NCBI database: ASM167528v1) (Kukutla et al., 2013; Lee et al., 2015; Matyi et al., 2013). The chromosomal size of E. miricola BM10 was 4.24 M bases, with 35.7% GC content, and 3,720 protein-coding genes. The chromosomal DNA size and GC content of E. miricola BM10 was similar to that of other strains. The E. miricola BM10 gene number was 3,873, which fell somewhere between the gene numbers of E. anophelis R26 and E. miricola GTC 862. The ANI of E. miricola BM10, as determined by pairwise comparison with E. anophelis R26, E. meningoseptica ATCC 13253, and E. miricola GTC 862, was 91.5%, 81.2%, and 94.3%, respectively. The ANI values of <95% suggested that E. miricola BM10 may be a new species in the genus Elizabethkingia (Kim et al., 2014). E. miricola BM10, a new species, is proposed as an intestinal symbiotic microorganism that is important for the low-temperature growth of termites. Studies on the diversity of intestinal symbiotic microbes in termites will be the basis for understanding the ecology of termites and various efforts to reduce the damage of termites (Kim and Chung, 2017; Lee et al., 2017). Additionally, the further studies on E. miricola BM10 will reveal low temperature gene expression mechanism that produces cellulase only at low temperature, absence of endoglucase in adaptation for symbiosis with termites, and enzymatic characteristics of the low temperature expressing β-glucosidas.
| Strain | E. miricola BM10 | E. anophelis R26 | E. meningoseptica ATCC 13253 | E. miricola GTC 862 |
|---|---|---|---|---|
| Size (Mb) | 4.2 | 4.0 | 3.8 | 4.3 |
| GC% | 35.7 | 35.4 | 35.6 | 35.8 |
| Protein | 3,720 | 3,687 | 3,370 | 4,006 |
| rRNA | 15 | - | - | 6 |
| tRNA | 53 | 39 | 45 | 46 |
| Other RNA | 1 | - | - | 6 |
| Genes | 3,873 | 3,726 | 3,415 | 4,064 |
| Assembly ID* | ASM95566v1 | ASM33181v1 | ASM40141v1 | ASM167528v1 |
| Reference | (Lee et al., 2015) | (Kukutla et al., 2013) | (Matyi et al., 2013) | - |
4. CONCLUSION
E. miricola BM10, a symbiotic gut bacteria of the termite Reticulitermes speratus KMT001, showed high activity of both cellobiohydrolase and β-glucosidase, but did not show any endo-β-1,4-glucanase activity, which is an enzyme required for initial cellulose degradation (Cho et al., 2010). E. miricola BM10 produced considerable β-glucosidase at 10°C (Lee et al., 2018). Based on its fatty acid composition, whole-genome sequence, and bacterial physiology, E. miricola BM10 appears to be distinct from the other three type-strains in the genus Elizabethkingia. It is therefore proposed that E. miricola BM10 is a new species in the genus Elizabethkingia.
