Original Article
Symbiotic Bacterial Flora Changes in Response to Low Temperature in Reticulitermes speratus KMT001
Dongmin Lee
2, Yeong-Suk Kim
2, Young-Kyoon Kim
2, Tae-Jong Kim
2,†
2Department of Forest Products and Biotechnology, College of Science and Technology, Kookmin University, 77 Jeongneung-ro, Seongbuk-gu, Seoul 02707, Republic of Korea
© The Korean Society of Wood Science & Technology.
Received: Jul 10, 2018; Accepted: Nov 12, 2018
Published Online: Nov 25, 2018
Abstract
Lower termites require symbiotic microbes in their gut. The microbial communities in the termites must adapt to the termite temperature. Reticulitermes speratus KMT001 from Bukhan Mountain in Seoul may require a special symbiotic microorganisms for growth in low temperature Korean habitat. A metagenomics analysis showed a dramatic change in the symbiotic bacterial flora in the gut of R. speratus KMT001 in response to low temperatures of 4°C or 10°C. Elusimicrobia, which are endosymbionts of flagellate protists, is the dominant phylum in the termite gut at ≥15°C but its population decreased drastically at low temperature. Four representative bacterial strains isolated from R. speratus KMT001 in a previous study produced maximum β-glucosidase levels within the temperature range of 10°C–30°C. Elizabethkingia sp. BM10 produced β-glucosidase specifically at 10°C. This strain supported the existence of symbiotic bacteria for the low temperature habitat of the termite. This identified bacterium will be a resource for studying low temperature adaptation of termites, studying the gene expression at low temperatures, and developing an industrial cellulase at low temperature.
Keywords: symbiotic bacteria; Reticulitermes speratus; low temperature; Elizabethkingia
1. INTRODUCTION
Termites damage wood structures and cause large economic losses annually (Ghaly and Edwards, 2011). Wood invaded by termites loses its structural integrity without any apparent change but the damage is usually irreversible (Peterson et al., 2006). Termites that have invaded wood cannot be eliminated without removing the infected wood. The best way to control termites is preventing invasion. Therefore, understanding the natural habitat and survival strategy of termites is critical to control termite damage (Kim and Chung, 2017; Mun and Nicholas, 2017).
Termites inhabit all continents of the world except Antarctica (Evans et al., 2013). It is quite interesting how termites can inhabit a variety of regions from tropical rainforests to frozen soil. Reticulitermes speratus is a termite species found in Korea (Cho et al., 2010b). It has been suggested that termites originally landed on the southern region of the Korean peninsula from Japan and migrated north (Park et al., 2006; Kim et al., 2012). Termites are currently found on Bukhan Mountain in Seoul, which is in the northern part of South Korea. Mean temperature in Korea during winter is close to 2°C (Jung et al., 2002), but termites survive and thrive the following spring.
Lower termites are considered as less evolved termites. Six out of seven termite families are lower termites. They obtain nutrients and energy mainly by digesting wood but they require support from symbiotic microbes in their gut (Brune and Ohkuma, 2011). These symbiotic microbes include protozoa (Cleveland, 1923), bacteria (Brune, 2014), and archaea (Shi et al., 2015). All symbiotic microbes and the termites themselves digest wood harmonically and synergistically (Brune, 2014). Disrupting this bacterial symbiosis with antibiotics significantly affects the normal physiology of termites (Rosengaus et al., 2011). As termites are small, their body temperature is greatly affected by the environment and the symbiotic microbes will also be exposed to the low temperature. It is unknown how symbiotic bacteria survive at low temperatures or how they maintain their essential support for survival of the termite.
The cellulose digestive system of termites using cellulase is complex (Lee et al., 2010). The termite digestive system produces endo-β-glucanase, which is an endotype cellulase, secreted in the mouth (Watanabe and Tokuda, 2010). However, lower termites require additional cellulases and many other enzymes from symbiotic microbes to fully digest wood (Brune, 2014). Lower termites cannot survive on wood alone without these symbiotic microbes, protozoa, bacteria, and archaea. In a previous study, 16 symbiotic bacteria were isolated from R. speratus KMT001 (Cho et al., 2010a). All of the strains had only cellobiohydrolase and β-glucosidase activities but no endo-β-glucanase activity. Considering that symbiotic bacteria have been isolated from the termite hindgut, we hypothesized that the loss of endo-β-glucanase activity was a consequence of symbiotic adaptation. The complete dependency of lower termites on symbiotic microbes for digesting wood may be due to diverse symbiotic biological functions (Brune, 2014; Peterson and Scharf, 2016). Considering the diverse habitats of termites and the requirements of symbiotic microbes, the symbiotic microbial population must change according to the termite’s habitat.
In this study, we investigated the change in the symbiotic bacteria population in the lower termite, R. speratus KMT001, from Bukhan Mountain according to a change in temperature, particularly low temperature. The results will provide clues as to how lower termites receive support from symbiotic microbes and how they survive low temperatures.
2. MATERIALS and METHODS
2.1. Purification of chromosomal DNA from symbiotic bacteria
Lower termites, Reticulitermes speratus KMT001 (Cho et al., 2010b), were collected from Bukhan Mountain, Seoul, Korea; 50 worker termites were grown at 4°C, 10°C, 15°C, 22°C, and 26°C for 14 days, and their guts were extracted with tweezers (Kim et al., 2010). The termite gut was extracted at the same temperature as the growing conditions for the sample to maintain the bacterial flora. The extracted gut was suspended in 250 μl of 50 mM EDTA (pH 8.0) and 50 μl of 10 g/l lysozyme and incubated at 37°C for 1 h. The supernatant was removed after 1 min centrifugation at 12,300 relative centrifugal force (RCF). Chromosomal DNA was purified with the MG Genomic DNA purification Kit (MGmed, Inc., Seoul, Korea). The AL buffer (300 μl) was added to the pellet and resuspended by gentle pipetting. The mixture was incubated at 80°C for 5 min. After cooling to room temperature, 1.5 μl RNase A solution was added, mixed five times by inversion, and incubated in a 37°C water bath for 1 h. After cooling to room temperature, 100 μl PP buffer was added, mixed by vigorous vortexing for 20 sec, and incubated in ice for 5 min. After centrifugation at 12,300 RCF for 3 min, the supernatant was transferred to a 1.5 ml Eppendorf tube containing 300 μl isopropanol. After centrifugation at 12,300 RCF for 1 min, the supernatant was removed, 300 μl of 70% ethanol was added at 4°C, and mixed by inversion several times. After centrifugation at 12,300 RCF for 1 min, the supernatant was removed again, and the remaining ethanol was evaporated. The dried chromosomal DNA was rehydrated with 100 μl distilled water in a 65°C water bath for 1 h. Two independent chromosomal DNA samples were prepared for each termite growth temperature.
2.2. Metagenomics analysis
A metagenomics analysis was performed by Macrogen Co. (Seoul, Korea). The libraries were prepared using polymerase chain reaction (PCR) according to the Rapid Library Preparation Method Manual (454 Life Sciences Corp., Branford, CT, USA). The libraries were quantified using the Picogreen assay (Ahn et al., 1996) and Victor3 (PerkinElmer, Inc., Waltham, MA, USA). Emulsion-based clonal amplification (emPCR amplification), corresponding to clonal amplification of the purified library, was carried out using the GS FLX Titanium MV emPCR Kit (454 Life Sciences Co.). Briefly, the library was immobilized on DNA capture beads. The library beads were added to a mixture of amplification mix and oil and shaken vigorously on a TissueLyser II (Qiagen Korea Ltd., Seoul, Korea) to create “micro-reactors” containing the amplification mix and a single bead. The emulsion was dispensed into a 96-well plate, and the PCR amplification program was run according to the manufacturer’s recommendations. A 20 ng aliquot of each sample DNA was used for a 50 μl PCR reaction. The 27F (5′-GAGTTTGATCMTGGCTCAG-3′) and 518R (5′-WTTACCGCGGCTGCTGG-3′) 16S universal primers were used to amplify the 16s rRNA gene. The FastStart High Fidelity PCR System (Roche Diagnostics, Seoul, Korea) was used for the PCR analysis under the following conditions: 94°C for 3 min, followed by 35 cycles of 94°C for 15 sec, 55°C for 45 sec, 72°C for 1 min, and a final elongation step at 72°C for 8 min. After PCR, the product was purified using AMPure beads (Beckman Coulter Korea Ltd., Seoul, Korea) and sequenced by next generation sequencing using the 454 Genome Sequencer-FLX plus (Roche Diagnostics). Each sample was loaded in one region of a 70 mm × 75 mm PicoTiter plate (454 Life Sciences Co.) fitted with an 8-lane gasket.
2.3. Selection of 16S rRNAs and taxonomic assignments
All sequence reads were compared to the Silva rRNA database. Sequence reads with a similar sequence and an E-value <0.01 were admitted as partial 16S rRNA sequences. Less than 1% was non-16S rRNA sequence reads. The taxonomic assignments of the sequenced reads were carried out using the Taxonomy Databases of National Center for Biotechnology Information. The five most similar sequences for each sequence read were found in the database using their bit score and E-value from the BLAST program. The Needleman-Winch global alignment algorithm was used to find the optimum alignment of the two sequences along their entire length. Pairwise global alignment was performed on selected candidate hits to identify the best aligned hit. The taxonomy of the sequence with the highest similarity was assigned to the sequence read. Taxonomy was assigned to species with >97% similarity, to genera with 94% similarity, to families with 90% similarity, to orders with 85% similarity, to classes with 80% similarity, and to phyla with 75% similarity.
2.4. Operational taxonomic unit (OTU) analysis for community richness
CD-HIT-OTU and Mothur software (Needleman and Wunsch, 1970) was used for clustering. The Shannon- Weaver diversity index and Simpson’s index were used to determine species diversity in the microbial communities.
2.5. β-glucosidase activity assay
β-glucosidase activity was measure according to a previous study (Inoue et al., 1997) with a few modifications. The bacterial strains from the previous study (Cho et al., 2010a) were cultured in YP-CMC media (0.8% peptone, 0.2% yeast extract, 0.5% potassium phosphate monobasic, 0.5% potassium phosphate dibasic, 2% carboxymethyl cellulose, and 0.025% antiform) for seven days with 200 rpm shaking at the indicated temperature. A 100 μl aliquot of the culture supernatant was mixed with 800 μl of 0.1 M sodium acetate buffer (pH 5.5) and 100 μl of 10 mM ρ-nitrophenyl-β-D-glucopyranoside, incubated at 50°C for 15 min, and the reaction was stopped by mixing 100 μl 2 M sodium carbonate. Absorbance was measured at a wavelength of 405 nm and the amount of ρ-nitrophenol produced was calculated using a ρ -nitrophenol standard curve. One unit of activity was defined as the amount of β-glucosidase activity that generated 1 μmol ρ-nitrophenol/min. Relative activity was calculated based on the highest activity of the strain among the tested temperatures.
2.6. Changes in growth at different temperatures
The strains were inoculated in 100 ml YP-CMC media and cultured at the indicated temperatures with 250 rpm shaking. Cell density was estimated by measuring absorbance at 600 nm at the indicated times. The growth curve was drawn as a semi-logarithmic plot, and the doubling times of the strains at each temperature were calculated based on this semi-logarithmic plot.
3. RESULTS and DISCUSSION
3.1. Change in gut bacterial population by temperature
The population studies of symbiotic bacteria according to changes in termite growing temperature identified different predominant phylogenetic bacterial groups (Supplementary Tables 1–6). At the class level, Elusimicrobia was the predominant population at 22°C and Gammaproteobacteria was predominant at 4°C (Supplementary Table 3). Enterobacteriales and Enterobacteriaceae were the predominant order and family of bacteria at 4°C, respectively (Supplementary Tables 4 and 5, respectively). Aestuariimicrobium, Citrobacter, Lactococcus, Serratia, and Treponema were the main genera at 4°C (Supplementary Table 6). All five major genera were representative examples of the four predominant phyla at 4°C.
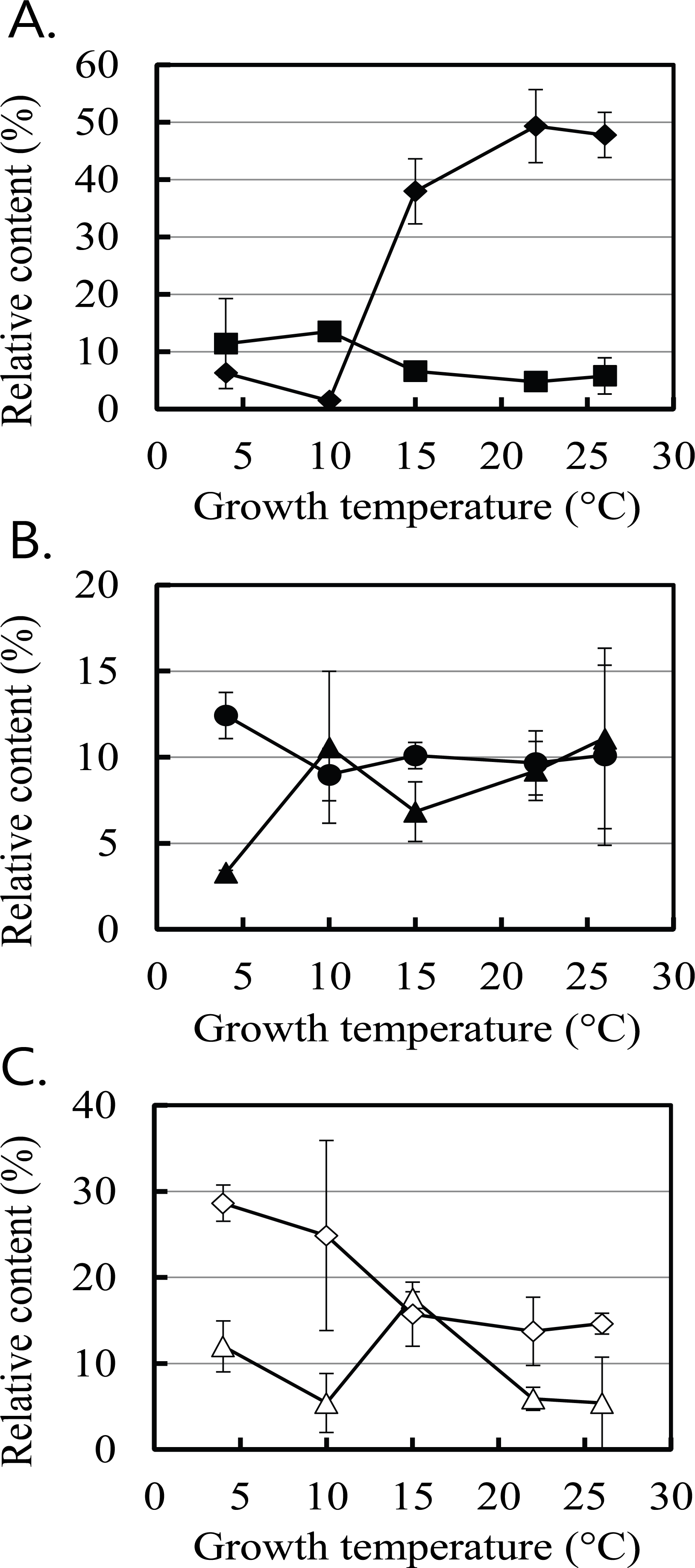
The changes in the six major populations at the phylum level with the unknown strains were selected from Supplementary Table 2 and drawn in Fig. 1. The proportions of Actinobacter, Firmicutes, Proteobacteria, and Spirochaetes increased at 4°C and 10°C, whereas those of Bacteroidetes and Elusimicrobia increased at 22°C and 26°C compared to 4°C. The salient phylum was Elusimicrobia. The mean proportions of the Elusimicrobia populations at 22°C and 26°C increased more than 12-fold from the mean proportions observed at 4°C and 10°C. The decrease in the proportion of many other phyla at 22°C and 26°C appeared to be due to the increase in the proportion of Elusimicrobia.
Fig. 1.
Changes in the proportions of six major symbiotic bacteria phyla in Reticulitermes speratus KMT001 based on temperature. The six major phyla were: Spirochaetes (■ in A), Elusimicrobia (◆ in A), Bacteroidetes (▲ in B), Actinobacteria (● in B), Proteobacteria (◇ in C), and Firmicutes (△ in C). The values are means of two independent experiments.
Download Original Figure
All phyla mentioned above were identified in previous studies (Ohkuma and Kudo, 1996; Hongoh et al., 2003; Yang et al., 2005). The termite Group I phylum has been identified as symbiotic bacteria in the hindgut of the lower termite Reticulitermes speratus (Ohkuma and Kudo, 1996) and renamed phylum Elusimicrobia (Geissinger et al., 2009). Elusimicrobia are endosymbionts of symbiotic flagellate protists in termites and the major bacteria among symbiotic bacteria in the termite gut (Yang et al., 2005). Fig. 1 confirms that the majority of the termite symbiotic bacteria were Elusimicrobia only at temperatures ≥15°C. Elusimicrobia was not the main phylum in the termite gut at 4°C and 10°C. Because Elusimicrobia is associated with protists (Yang et al., 2005), it was expected that the change in the Elusimicrobia population would reflect the population change in protists. Growth of protists is significantly inhibited at temperatures <15°C (Rose and Caron, 2007), supporting the hypothesis that the decrease in the Elusimicrobia population was due to the reduced number of host flagellate protists at 4°C and 10°C. Growth of symbiotic protists and endosymbiotic Elusimicrobia may be inhibited when temperature of the termites is low, such as 4°C and 10°C. The drastic decrease in the Elusimicrobia population may be the main contributor to the increase in the populations of other bacteria at 4°C and 10°C. Notably, the unidentified phylum of bacteria in Supplementary Table 2 (shown as ‘The rest’) was >25% of the total population at 4°C and 10°C. Because most studies on symbiotic bacteria were performed at room temperature or higher (Ohkuma and Kudo, 1996; Hongoh et al., 2003; Yang et al., 2005; Herlemann et al., 2007), many of the symbiotic bacteria found at low temperatures in this study may not have been known.
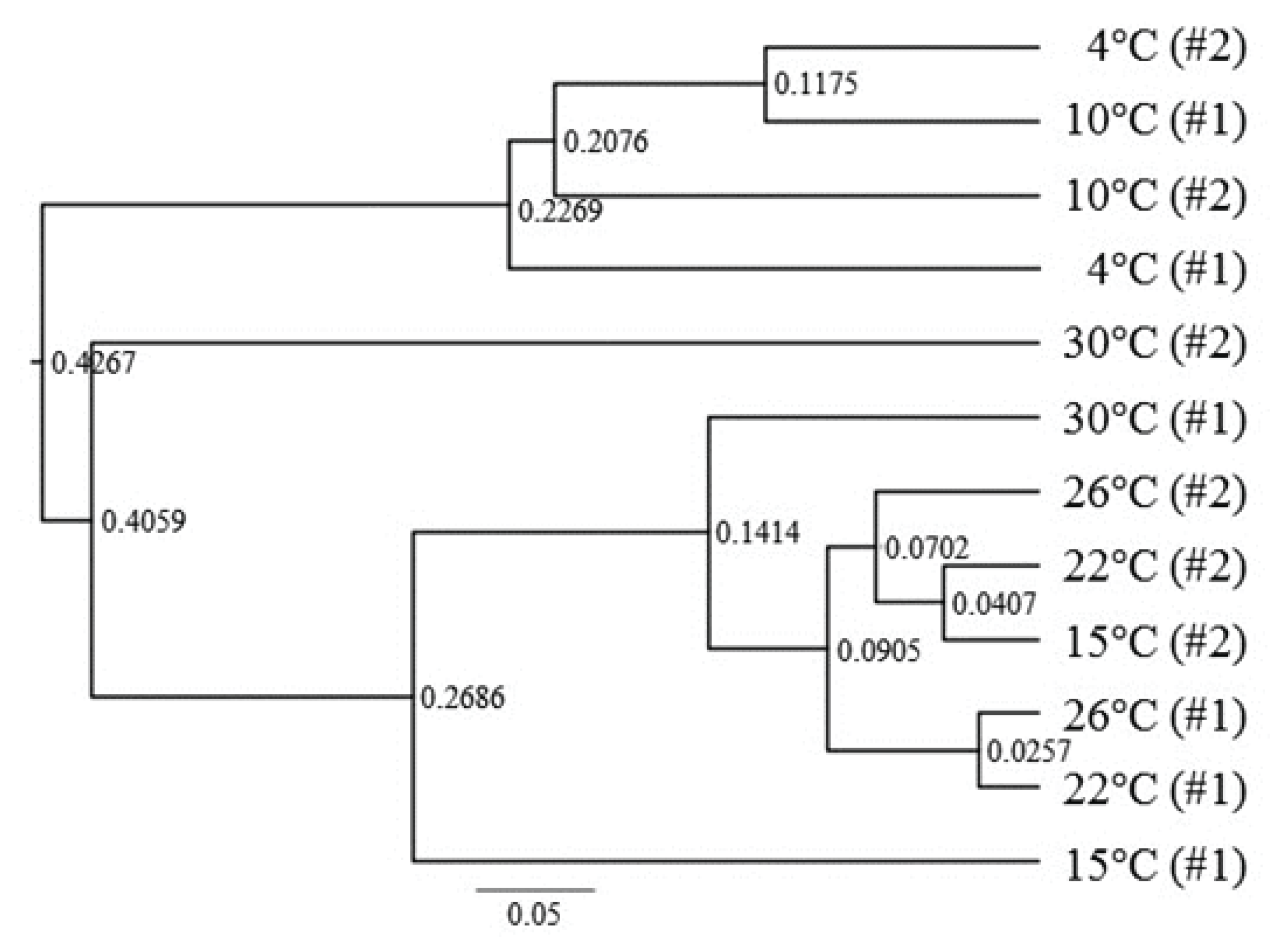
The beta diversity results in Fig. 2 show that the symbiotic bacterial populations in the termite gut at 4°C and 10°C were quite different from those observed at ≥15°C. Therefore, the beta diversity analysis of the metagenomics results supports the population analysis at the phylum level in Fig. 1.
Fig. 2.
Operational taxonomic unit (OTU) analysis for community richness. The symbiotic bacteria flora samples in the termites at each temperature were taken independently and are shown as #1 and #2 for each sample. Numbers on the nodes at branch separation represent node depths. The ruler bar at the bottom indicates 0.05 relative depth.
Download Original Figure
3.2. Symbiotic bacteria supporting termites at low temperature
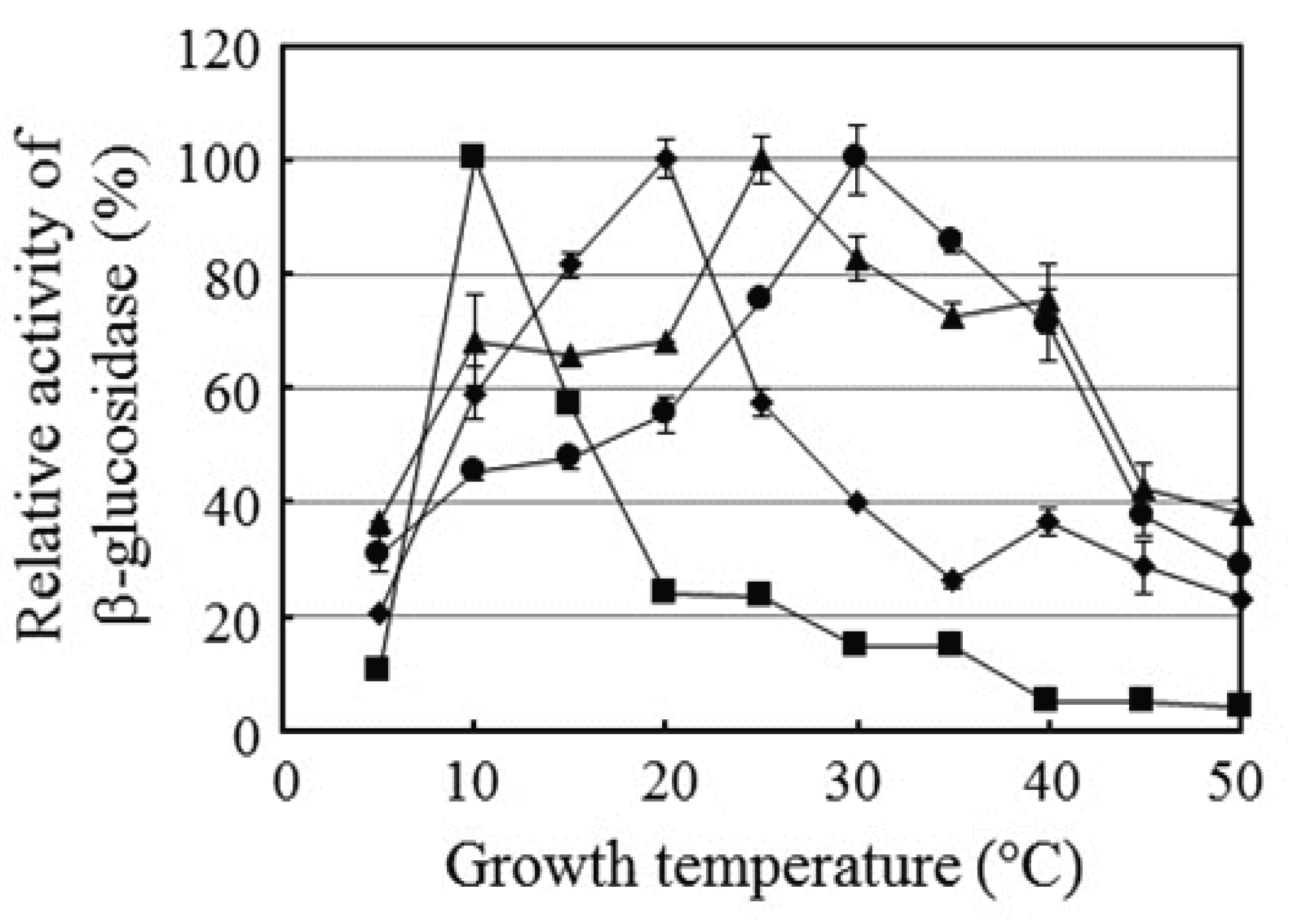
Lower termites growing at low temperatures still need support from symbiotic microbes to survive. The Elusimicrobia populations at 4°C and 10°C decreased dramatically, and it is expected that the number of protists also decreased. Therefore, termites require symbiotic support from other microbes. In a previous study, 16 bacteria were isolated from the lower termite R. speratus KMT001 (Cho et al., 2010a). All produced cellobiohydrolase and β-glucosidase but not endo-β-glucanase. In this study, we tested the temperature effect on β-glucosidase production by the isolated strains. The 16 strains tested showed the activity of on β-glucosidase at various temperature and the results of four representative strains of them were shown in Fig. 3. Elizabethkingia sp. BM10, Bacillus sp. NT4, Serratia sp. PT1B, and Serratia sp. NT3 had maximum β-glucosidase activity when grown at 10°C, 20°C, 25°C, and 30°C, respectively, indicating that R. speratus KMT001 has broad bacterial diversity that produced β-glucosidase within the temperature range of the termite habitat, including 10°C. The interesting observation was β-glucosidase production by Elizabethkingia sp. BM10. An increase in temperature by 5°C from the temperature that yielded the highest β-glucosidase activity reduced β-glucosidase production by more than 40%, suggesting that Elizabethkingia sp. BM10 is specialized to produce β-glucosidase at 10°C and that this strain supports cellulose digestion in the termite gut at low temperatures.
Fig. 3.
Effect of temperature on β-glucosidase production by four symbiotic bacteria from Reticulitermes speratus KMT001. Relative β-glucosidase activity of culture media was calculated based on the maximum activity of the tested strain after a 7-day culture. The four strains were Elizabethkingia sp. BM10 (■), Bacillus sp. NT4 (◆), Serratia sp. PT1B (▲), and Serratia sp. NT3 (●). Values are means of three independent experiments.
Download Original Figure
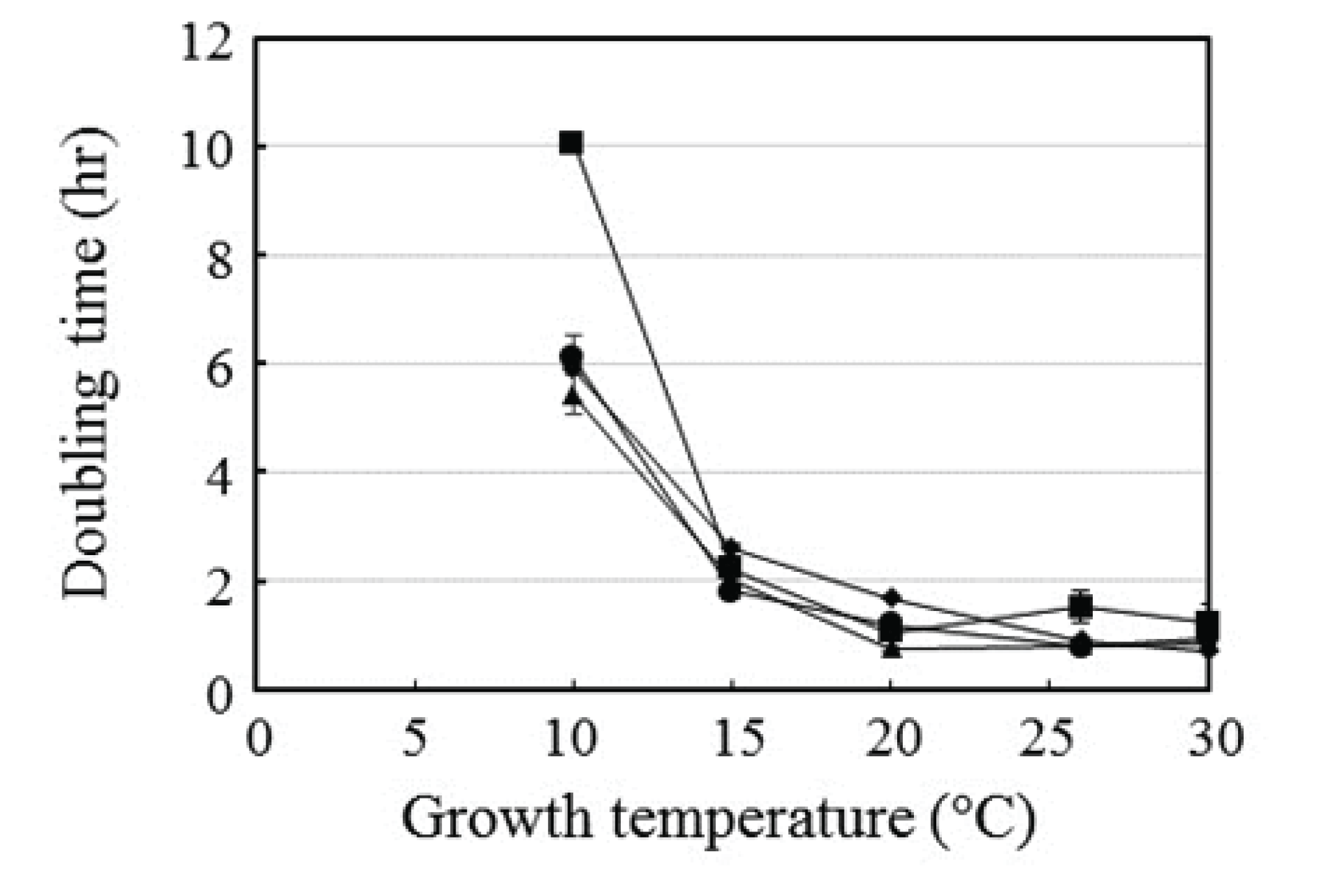
The growth rates of four representative isolated symbiotic bacteria were tested (Fig. 4). The doubling times of the strains were similar at most of the temperatures tested, except 10°C, at which the doubling time of Elizabethkingia sp. BM10 was almost twice that of the other three strains. This finding indicates that the growth rate of Elizabethkingia sp. BM10 was lower than that of the other strains. The slow growth rate of Elizabethkingia sp. BM10 can be explained by the strong β-glucosidase production at 10°C (Kafri et al., 2016).
Fig. 4.
Effect of temperature on the doubling time of the four strains: Elizabethkingia sp. BM10 (■), Bacillus sp. NT4 (◆), Serratia sp. PT1B (▲), and Serratia sp. NT3 (●). The growth curve was observed at the indicated temperature, and the doubling times were calculated during the exponential phase. Values are means of two independent experiments, except that for Elizabethkingia sp. BM10 whose mean value was from three independent experiments.
Download Original Figure
This study showed a dramatic change in the symbiotic bacterial flora of R. speratus KMT001 at low temperatures of 4°C and 10°C. More than 25% of the bacteria in the metagenomics analysis were unknown strains, suggesting that more intense studies on symbiotic bacteria of termites are required at low temperature. Elizabethkingia sp. BM10 produced maximal β-glucosidase levels at 10°C, which is an unusual temperature for β-glucosidase production. We suggest that Elizabethkingia sp. BM10 is one of the strains supporting cellulose digestion in termites at low temperature.
4. CONCLUSION
Termites are harmful insects causing large economic losses and also models for the biological degradation system of woods. Lower termites require a support of symbiotic microbes in the digestion. However, their symbiosis at low temperature has not been studied well. In this study, we showed a significant change of symbiotic bacterial flora by low temperature and present one example of isolated bacteria strain supporting the symbiosis at low temperature. This study provides clues how termites can survive with symbiotic supports and demonstrates the symbiotic adaptation of bacterial flora in the termite guts at low temperature.
ACKNOWLEDGMENT
This study was carried out with the support of ‘R&D Program for Forest Science Technology (Project No. 2013070E10-1819-AA03)’ provided by Korea Forest Service (Korea Forestry Promotion Institute).
SUPPLEMENTARY MATERIAL
Supplemental Table 1.
Metagenomics analysis of the temperature effect on the symbiotic bacterial population of Reticulitermes speratus KMT001 at the superkingdom level.
|
Superkingdom |
Relative content (%) and standard deviation in parenthesis |
|
4°C |
10°C |
15°C |
22°C |
26°C |
|
Bacteria |
100 (±0.00) |
100 (±0.00) |
100 (±0.00) |
100 (±0.00) |
100 (±0.00) |
|
The rest |
0 (±0.00) |
0 (±0.00) |
0 (±0.00) |
0 (±0.00) |
0 (±0.00) |
Download Excel Table
Supplemental Table 2.
Metagenomics analysis of the temperature effect on the symbiotic bacterial population of Reticulitermes speratus KMT001 at the phylum level.
|
Phylum |
Relative content (%) and standard deviation in parenthesis |
|
4°C |
10°C |
15°C |
22°C |
26°C |
|
Acidobacteria |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.23 (±0.33) |
0.08 (±0.11) |
|
Actinobacteria |
11.39 (±7.85) |
13.52 (±1.47) |
6.58 (±1.54) |
4.75 (±0.73) |
5.76 (±3.16) |
|
Bacteroidetes |
3.30 (±0.12) |
10.58 (±4.41) |
6.83 (±1.73) |
9.20 (±1.71) |
11.09 (±5.24) |
|
Chlorobi |
0.06 (±0.08) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.04) |
0.00 (±0.00) |
|
Cyanobacteria |
0.00 (±0.00) |
0.01 (±0.01) |
0.10 (±0.06) |
0.06 (±0.07) |
0.00 (±0.00) |
|
Deinococcus-Thermus |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.04) |
0.30 (±0.31) |
0.00 (±0.00) |
|
Elusimicrobia |
6.26 (±0.70) |
1.47 (±0.40) |
37.93 (±5.68) |
49.31 (±6.38) |
47.76 (±3.94) |
|
Firmicutes |
12.43 (±1.34) |
8.99 (±1.51) |
10.09 (±0.77) |
9.67 (±1.86) |
10.11 (±5.23) |
|
Fusobacteria |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.02) |
0.00 (±0.00) |
|
Planctomycetes |
0.14 (±0.08) |
0.18 (±0.01) |
0.04 (±0.06) |
0.11 (±0.07) |
0.03 (±0.04) |
|
Proteobacteria |
28.63 (±2.10) |
24.87 (±11.05) |
15.73 (±3.71) |
13.75 (±3.97) |
14.64 (±1.21) |
|
Spirochaetes |
11.99 (±2.97) |
5.41 (±3.42) |
17.58 (±0.75) |
5.90 (±1.35) |
5.42 (±5.33) |
|
Synergistetes |
0.44 (±0.31) |
0.65 (±0.30) |
0.88 (±0.03) |
0.84 (±0.16) |
0.31 (±0.21) |
|
Tenericutes |
0.46 (±0.10) |
0.27 (±0.00) |
0.36 (±0.13) |
0.80 (±0.97) |
0.26 (±0.22) |
|
Verrucomicrobia |
0.22 (±0.15) |
0.07 (±0.10) |
0.21 (±0.21) |
0.17 (±0.17) |
0.35 (±0.25) |
|
The rest |
24.68 (±4.46) |
33.97 (±2.38) |
3.63 (±1.20) |
4.84 (±1.19) |
4.18 (±0.46) |
Download Excel Table
Supplemental Table 3.
Metagenomics analysis of the temperature effect on the symbiotic bacterial population of Reticulitermes speratus KMT001 at the class level.
|
Class |
Relative content (%) and standard deviation in parenthesis |
|
4°C |
10°C |
15°C |
22°C |
26°C |
|
Acidobacteria |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.23 (±0.33) |
0.08 (±0.11) |
|
Actinobacteria |
11.39 (±7.85) |
13.52 (±1.47) |
6.58 (±1.54) |
4.75 (±0.73) |
5.76 (±3.16) |
|
Alphaproteobacteria |
4.77 (±0.46) |
2.36 (±1.80) |
10.56 (±0.95) |
9.70 (±4.96) |
8.91 (±0.82) |
|
Bacilli |
10.47 (±2.03) |
4.12 (±0.32) |
5.93 (±1.05) |
7.15 (±1.38) |
7.24 (±3.80) |
|
Bacteroidia |
3.29 (±0.11) |
10.39 (±4.32) |
6.56 (±1.59) |
8.91 (±1.31) |
10.80 (±5.50) |
|
Betaproteobacteria |
1.38 (±0.66) |
5.99 (±6.23) |
4.06 (±2.38) |
2.07 (±1.05) |
1.01 (±0.87) |
|
Chlorobia |
0.06 (±0.08) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.04) |
0.00 (±0.00) |
|
Clostridia |
1.88 (±0.74) |
4.85 (±1.19) |
3.94 (±0.32) |
2.49 (±0.46) |
2.84 (±1.47) |
|
Deltaproteobacteria |
0.29 (±0.10) |
0.08 (±0.00) |
0.73 (±0.48) |
0.41 (±0.18) |
0.07 (±0.07) |
|
Elusimicrobia |
6.26 (±0.70) |
1.47 (±0.40) |
37.93 (±5.68) |
49.31 (±6.38) |
47.76 (±3.94) |
|
Epsilonproteobacteria |
0.19 (±0.17) |
0.28 (±0.16) |
0.10 (±0.08) |
0.03 (±0.04) |
0.00 (±0.00) |
|
Erysipelotrichi |
0.08 (±0.05) |
0.02 (±0.00) |
0.22 (±0.04) |
0.03 (±0.02) |
0.03 (±0.04) |
|
Flavobacteria |
0.01 (±0.02) |
0.19 (±0.09) |
0.27 (±0.15) |
0.29 (±0.40) |
0.29 (±0.26) |
|
Fusobacteria |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.02) |
0.00 (±0.00) |
|
Gammaproteobacteria |
22.00 (±0.92) |
16.16 (±18.91) |
0.27 (±0.19) |
1.54 (±0.27) |
4.64 (±1.19) |
|
Mollicutes |
0.46 (±0.10) |
0.27 (±0.00) |
0.36 (±0.13) |
0.80 (±0.97) |
0.26 (±0.22) |
|
Opitutae |
0.22 (±0.15) |
0.07 (±0.10) |
0.21 (±0.21) |
0.17 (±0.17) |
0.35 (±0.25) |
|
Planctomycetacia |
0.14 (±0.08) |
0.17 (±0.03) |
0.04 (±0.05) |
0.09 (±0.09) |
0.03 (±0.04) |
|
Sphingobacteria |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Spirochaetes |
11.99 (±2.97) |
5.41 (±3.42) |
17.58 (±0.75) |
5.90 (±1.35) |
5.42 (±5.33) |
|
Synergistia |
0.44 (±0.31) |
0.65 (±0.30) |
0.88 (±0.30) |
0.84 (±0.16) |
0.31 (±0.21) |
|
The rest |
24.68 (±4.46) |
33.99 (±2.41) |
3.77 (±1.30) |
5.22 (±0.83) |
4.18 (±0.46) |
Download Excel Table
Supplemental Table 4.
Metagenomics analysis of the temperature effect on the symbiotic bacterial population of Reticulitermes speratus KMT001 at the order level.
|
Order |
Relative content (%) and standard deviation in parenthesis |
|
4°C |
10°C |
15°C |
22°C |
26°C |
|
Acidobacteriales |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.23 (±0.33) |
0.08 (±0.11) |
|
Actinomycetales |
11.21 (±7.85) |
13.43 (±1.49) |
6.49 (±1.51) |
4.69 (±0.75) |
5.69 (±3.08) |
|
Bacillales |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.12 (±0.03) |
0.43 (±0.61) |
|
Bacteroidales |
3.29 (±0.11) |
10.39 (±4.32) |
6.56 (±1.59) |
8.91 (±1.31) |
10.80 (±5.50) |
|
Burkholderiales |
0.41 (±0.51) |
2.75 (±3.18) |
0.14 (±0.05) |
0.13 (±0.19) |
0.36 (±0.30) |
|
Campylobacterales |
0.19 (±0.17) |
0.28 (±0.16) |
0.10 (±0.08) |
0.03 (±0.04) |
0.00 (±0.00) |
|
Caulobacterales |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.10 (±0.01) |
0.00 (±0.00) |
|
Chlorobiales |
0.06 (±0.08) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.04) |
0.00 (±0.00) |
|
Clostridiales |
1.88 (±0.74) |
4.85 (±1.19) |
3.94 (±0.32) |
2.49 (±0.46) |
2.84 (±1.47) |
|
Coriobacteriales |
0.18 (±0.00) |
0.09 (±0.02) |
0.08 (±0.03) |
0.07 (±0.02) |
0.07 (±0.08) |
|
Desulfovibrionales |
0.15 (±0.08) |
0.05 (±0.04) |
0.61 (±0.34) |
0.36 (±0.20) |
0.07 (±0.08) |
|
Elusimicrobiales |
0.00 (±0.00) |
0.04 (±0.06) |
0.03 (±0.04) |
0.01 (±0.01) |
0.00 (±0.00) |
|
Enterobacteriales |
22.00 (±0.92) |
15.79 (±19.23) |
0.03 (±0.04) |
1.45 (±0.31) |
4.20 (±1.78) |
|
Erysipelotrichales |
0.08 (±0.05) |
0.02 (±0.00) |
0.22 (±0.04) |
0.03 (±0.02) |
0.03 (±0.04) |
|
Flavobacteriales |
0.01 (±0.02) |
0.19 (±0.09) |
0.27 (±0.15) |
0.29 (±0.40) |
0.29 (±0.26) |
|
Fusobacteriales |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.02) |
0.00 (±0.00) |
|
Lactobacillales |
10.47 (±2.03) |
4.12 (±0.32) |
5.93 (±1.05) |
7.03 (±1.41) |
6.81 (±3.19) |
|
Legionellales |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.08 (±0.11) |
|
Mycoplasmatales |
0.46 (±0.10) |
0.27 (±0.00) |
0.36 (±0.13) |
0.80 (±0.97) |
0.26 (±0.22) |
|
Neisseriales |
0.08 (±0.05) |
2.55 (±3.61) |
0.02 (±0.01) |
0.19 (±0.14) |
0.03 (±0.04) |
|
Planctomycetales |
0.14 (±0.08) |
0.17 (±0.03) |
0.04 (±0.05) |
0.09 (±0.09) |
0.03 (±0.04) |
|
Pseudomonadales |
0.00 (±0.00) |
0.35 (±0.28) |
0.19 (±0.08) |
0.02 (±0.03) |
0.15 (±0.21) |
|
Rhizobiales |
0.00 (±0.00) |
0.04 (±0.06) |
0.07 (±0.10) |
0.10 (±0.12) |
0.17 (±0.03) |
|
Rhodocyclales |
0.90 (±0.20) |
0.69 (±0.56) |
3.89 (±2.43) |
1.75 (±1.10) |
0.63 (±0.61) |
|
Rhodospirillales |
0.25 (±0.13) |
0.16 (±0.01) |
0.31 (±0.10) |
0.57 (±0.63) |
0.15 (±0.02) |
|
Rickettsiales |
4.52 (±0.59) |
2.02 (±1.59) |
10.18 (±0.94) |
8.89 (±4.39) |
8.48 (±1.01) |
|
Sphingobacteriales |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Sphingomonadales |
0.00 (±0.00) |
0.14 (±0.16) |
0.00 (±0.00) |
0.03 (±0.04) |
0.12 (±0.17) |
|
Spirochaetales |
11.99 (±2.97) |
5.41 (±3.42) |
17.58 (±0.75) |
5.90 (±1.35) |
5.42 (±5.33) |
|
Synergistales |
0.44 (±0.31) |
0.65 (±0.30) |
0.88 (±0.30) |
0.84 (±0.16) |
0.31 (±0.21) |
|
Thermales |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.04) |
0.30 (±0.31) |
0.00 (±0.00) |
|
Xanthomonadales |
0.00 (±0.00) |
0.02 (±0.03) |
0.05 (±0.07) |
0.07 (±0.00) |
0.21 (±0.27) |
|
The rest |
31.30 (±5.32) |
35.53 (±2.89) |
41.97 (±7.05) |
54.45 (±7.69) |
52.30 (±3.25) |
Download Excel Table
Supplemental Table 5.
Metagenomics analysis of the temperature effect on the symbiotic bacterial population of Reticulitermes speratus KMT001 at the family level.
|
Family |
Relative content (%) and standard deviation in parenthesis |
|
4°C |
10°C |
15°C |
22°C |
26°C |
|
Acetobacteraceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.08 (±0.11) |
0.05 (±0.07) |
|
Acidobacteriaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.23 (±0.33) |
0.08 (±0.11) |
|
Actinomycetaceae |
0.00 (±0.00) |
0.05 (±0.07) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Alcaligenaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.06) |
|
Alicyclobacillaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.03) |
0.00 (±0.00) |
|
Aurantimonadaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.09 (±0.07) |
|
Bacillaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.19 (±0.26) |
|
Bradyrhizobiaceae |
0.00 (±0.00) |
0.04 (±0.06) |
0.02 (±0.02) |
0.08 (±0.09) |
0.04 (±0.05) |
|
Brucellaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.03) |
|
Burkholderiaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.06) |
0.13 (±0.19) |
0.09 (±0.08) |
|
Caulobacteraceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.10 (±0.01) |
0.00 (±0.00) |
|
Clostridiaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 ±0.00) |
0.00 (±0.00) |
|
Comamonadaceae |
0.41 (±0.51) |
2.75 (±3.18) |
0.06 (±0.06) |
0.00 (±0.00) |
0.23 (±0.32) |
|
Coriobacteriaceae |
0.18 (±0.00) |
0.09 (±0.02) |
0.08 (±0.03) |
0.07 (±0.02) |
0.07 (±0.08) |
|
Corynebacteriaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.02) |
0.01 (±0.02) |
2.00 (±2.84) |
|
Coxiellaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.08 (±0.11) |
|
Desulfovibrionaceae |
0.15 (±0.08) |
0.05 (±0.04) |
0.61 (±0.34) |
0.36 (±0.20) |
0.07 (±0.08) |
|
Dietziaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Elusimicrobiaceae |
0.00 (±0.00) |
0.04 (±0.06) |
0.03 (±0.04) |
0.01 (±0.01) |
0.00 (±0.00) |
|
Enterobacteriaceae |
22.00 (±0.92) |
15.79 (±19.23) |
0.03 (±0.04) |
1.45 (±0.31) |
4.20 (±1.78) |
|
Enterococcaceae |
0.35 (±0.01) |
0.25 (±0.09) |
0.45 (±0.19) |
0.00 (±0.00) |
0.02 (±0.03) |
|
Erysipelotrichaceae |
0.08 (±0.05) |
0.02 (±0.00) |
0.22 (±0.04) |
0.03 (±0.02) |
0.03 (±0.04) |
|
Erythrobacteraceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Eubacteriaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.03) |
0.01 (±0.01) |
|
Flavobacteriaceae |
0.01 (±0.02) |
0.19 (±0.09) |
0.27 (±0.15) |
0.29 (±0.40) |
0.29 (±0.26) |
|
Fusobacteriaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.02) |
0.00 (±0.00) |
|
Helicobacteraceae |
0.19 (±0.17) |
0.28 (±0.16) |
0.10 (±0.08) |
0.03 (±0.04) |
0.00 (±0.00) |
|
Lachnospiraceae |
0.54 (±0.21) |
0.72 (±0.23) |
0.34 (±0.04) |
0.82 (±0.07) |
1.39 (±1.50) |
|
Lactobacillaceae |
1.44 (±1.23) |
1.18 (±0.99) |
0.06 (±0.09) |
0.62 (±0.02) |
1.07 (±1.47) |
|
Leuconostocaceae |
0.00 (±0.00) |
0.17 (±0.21) |
0.01 (±0.01) |
0.96 (±0.97) |
1.04 (±1.37) |
|
Methylobacteriaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.05 (±0.07) |
0.02 (±0.03) |
0.02 (±0.03) |
|
Methylocystaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Microbacteriaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.81 (±1.15) |
|
Moraxellaceae |
0.00 (±0.00) |
0.35 (±0.28) |
0.09 (±0.06) |
0.02 (±0.03) |
0.15 (±0.21) |
|
Mycobacteriaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.03) |
0.00 (±0.00) |
|
Mycoplasmataceae |
0.46 (±0.10) |
0.27 (±0.00) |
0.36 (±0.13) |
0.80 (±0.97) |
0.26 (±0.22) |
|
Neisseriaceae |
0.08 (±0.05) |
2.55 (±3.61) |
0.02 (±0.01) |
0.19 (±0.14) |
0.03 (±0.04) |
|
Opitutaceae |
0.07 (±0.10) |
0.01 (±0.01) |
0.01 (±0.01) |
0.02 (±0.03) |
0.00 (±0.00) |
|
Oxalobacteraceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.05) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Paenibacillaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.08 (±0.11) |
|
Peptococcaceae |
0.02 (±0.03) |
0.03 (±0.04) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Peptostreptococcaceae |
0.01 (±0.02) |
0.13 (±0.18) |
0.00 (±0.00) |
0.00 (±0.00) |
0.09 (±0.13) |
|
Planctomycetaceae |
0.14 (±0.08) |
0.17 (±0.03) |
0.04 (±0.05) |
0.09 (±0.09) |
0.03 (±0.04) |
|
Planococcaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.10 (±0.15) |
|
Porphyromonadaceae |
3.10 (±0.02) |
8.92 (±4.02) |
4.00 (±1.14) |
7.01 (±1.76) |
10.10 (±5.08) |
|
Propionibacteriaceae |
11.21 (±7.85) |
13.38 (±1.56) |
6.48 (±1.53) |
4.65 (±0.76) |
2.84 (±0.85) |
|
Pseudomonadaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.10 (±0.14) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Rhodocyclaceae |
0.90 (±0.20) |
0.69 (±0.56) |
3.89 (±2.43) |
1.75 (±1.10) |
0.63 (±0.61) |
|
Rhodospirillaceae |
0.25 (±0.13) |
0.16 (±0.01) |
0.31 (±0.10) |
0.49 (±0.52) |
0.10 (±0.09) |
|
Rikenellaceae |
0.07 (±0.00) |
0.36 (±0.12) |
0.28 (±0.08) |
0.22 (±0.02) |
0.18 (±0.18) |
|
Ruminococcaceae |
0.51 (±0.34) |
1.41 (±0.48) |
0.96 (±0.14) |
0.84 (±0.15) |
0.63 (±0.15) |
|
Sanguibacteraceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.05) |
|
Sphingomonadaceae |
0.00 (±0.00) |
0.05 (±0.07) |
0.00 (±0.00) |
0.03 (±0.04) |
0.12 (±0.17) |
|
Spirochaetaceae |
11.99 (±2.97) |
5.41 (±3.42) |
17.58 (±0.75) |
5.90 (±1.35) |
5.42 (±5.33) |
|
Sporolactobacillaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.06 (±0.06) |
0.01 (±0.02) |
|
Staphylococcaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.06) |
0.00 (±0.00) |
|
Streptococcaceae |
8.49 (±0.69) |
2.34 (±1.25) |
5.41 (±0.77) |
5.45 (±0.47) |
4.67 (±0.31) |
|
Synergistaceae |
0.44 (±0.31) |
0.65 (±0.30) |
0.88 (±0.30) |
0.84 (±0.16) |
0.31 (±0.21) |
|
Thermaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.04) |
0.30 (±0.31) |
0.00 (±0.00) |
|
Thermoactinomycetaceae |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.05 (±0.07) |
|
Veillonellaceae |
0.00 (±0.00) |
0.05 (±0.07) |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.05) |
|
Xanthomonadaceae |
0.00 (±0.00) |
0.02 (±0.03) |
0.05 (±0.07) |
0.07 (±0.00) |
0.21 (±0.27) |
|
The rest |
36.92 (±4.73) |
41.41 (±5.01) |
57.07 (±5.50) |
65.82 (±3.47) |
61.98 (±2.31) |
Download Excel Table
Supplemental Table 6.
Metagenomics analysis of the temperature effect on the symbiotic bacterial population of Reticulitermes speratus KMT001 at the genus level.
|
Genus |
Relative content (%) and standard deviation in parenthesis |
|
4°C |
10°C |
15°C |
22°C |
26°C |
|
Achromobacter
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.06) |
|
Acidiphilium
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.06) |
0.05 (±0.07) |
|
Acidisoma
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.06) |
0.00 (±0.00) |
|
Acidovorax
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.04) |
|
Acinetobacter
|
0.00 (±0.00) |
0.35 (±0.28) |
0.09 (±0.06) |
0.02 (±0.03) |
0.15 (±0.21) |
|
Actinomyces
|
0.00 (±0.00) |
0.05 (±0.07) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Aestuariimicrobium
|
11.17 (±7.92) |
13.20 (±1.34) |
6.40 (±1.42) |
4.33 (±0.57) |
2.56 (±0.85) |
|
Afipia
|
0.00 (±0.00) |
0.04 (±0.06) |
0.02 (±0.02) |
0.08 (±0.09) |
0.04 (±0.05) |
|
Alicyclobacillus
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.03) |
0.00 (±0.00) |
|
Alistipes
|
0.02 (±0.03) |
0.03 (±0.04) |
0.14 (±0.09) |
0.05 (±0.01) |
0.03 (±0.01) |
|
Anaerofustis
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.03) |
0.01 (±0.01) |
|
Anaerotruncus
|
0.00 (±0.00) |
0.05 (±0.07) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Bacillus
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.19 (±0.26) |
|
Brevinema
|
0.20 (±0.07) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Brevundimonas
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.05 (±0.07) |
0.00 (±0.00) |
|
Burkholderia
|
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.06) |
0.13 (±0.19) |
0.06 (±0.05) |
|
Candidatus Captivus
|
2.97 (±0.22) |
1.35 (±1.08) |
6.33 (±1.81) |
5.47 (±3.24) |
6.48 (±0.57) |
|
Candidatus Chloracidobacterium
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Candidatus Hepatincola
|
1.51 (±0.36) |
0.67 (±0.51) |
3.80 (±0.79) |
3.35 (±1.24) |
1.92 (±0.55) |
|
Candidatus Odyssella
|
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.02) |
0.06 (±0.08) |
0.00 (±0.00) |
|
Candidatus Symbiothrix
|
1.93 (±0.01) |
3.52 (±0.99) |
2.77 (±0.71) |
3.40 (±1.36) |
4.41 (±3.29) |
|
Candidatus Tammella
|
0.02 (±0.03) |
0.20 (±0.07) |
0.13 (±0.01) |
0.24 (±0.09) |
0.05 (±0.02) |
|
Chitinophaga
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Chryseobacterium
|
0.00 (±0.00) |
0.03 (±0.04) |
0.01 (±0.01) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Citrobacter
|
11.30 (±10.19) |
0.78 (±0.78) |
0.01 (±0.01) |
0.19 (±0.23) |
1.51 (±0.97) |
|
Cloacibacterium
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.16 (±0.23) |
|
Clostridium
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Comamonas
|
0.41 (±0.51) |
2.75 (±3.18) |
0.06 (±0.06) |
0.00 (±0.00) |
0.20 (±0.28) |
|
Corynebacterium
|
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.02) |
0.01 (±0.02) |
2.00 (±2.84) |
|
Cronobacter
|
0.33 (±0.04) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Cupriavidus
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.03) |
|
Daeguia
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.03) |
|
Delftia
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Desulfovibrio
|
0.15 (±0.08) |
0.05 (±0.04) |
0.61 (±0.34) |
0.36 (±0.20) |
0.07 (±0.08) |
|
Dietzia
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Dyella
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Dysgonomonas
|
0.08 (±0.01) |
2.70 (±2.20) |
0.34 (±0.02) |
0.81 (±0.18) |
2.19 (±0.57) |
|
Edaphobacter
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.06 (±0.08) |
0.00 (±0.00) |
|
Elizabethkingia
|
0.00 (±0.00) |
0.06 (±0.09) |
0.01 (±0.01) |
0.10 (±0.13) |
0.08 (±0.04) |
|
Elusimicrobium
|
0.00 (±0.00) |
0.04 (±0.06) |
0.03 (±0.04) |
0.01 (±0.01) |
0.00 (±0.00) |
|
Enterobacter
|
0.04 (±0.06) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Enterococcus
|
0.35 (±0.01) |
0.25 (±0.09) |
0.45 (±0.19) |
0.00 (±0.00) |
0.02 (±0.03) |
|
Ethanoligenens
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.03) |
|
Faecalibacterium
|
0.00 (±0.00) |
0.06 (±0.09) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Fusobacterium
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.02) |
0.00 (±0.00) |
|
Kiloniella
|
0.00 (±0.00) |
0.08 (±0.09) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Lactobacillus
|
1.44 (±1.23) |
1.18 (±0.99) |
0.06 (±0.09) |
0.62 (±0.02) |
1.07 (±1.47) |
|
Lactococcus
|
6.48 (±0.04) |
0.73 (±0.35) |
3.92 (±0.65) |
1.27 (±1.28) |
1.29 (±1.60) |
|
Leuconostoc
|
0.00 (±0.00) |
0.17 (±0.21) |
0.01 (±0.01) |
0.23 (±0.06) |
1.04 (±1.37) |
|
Lysinibacillus
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Methylobacterium
|
0.00 (±0.00) |
0.00 (±0.00) |
0.05 (±0.07) |
0.02 (±0.03) |
0.02 (±0.03) |
|
Microbacterium
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.37 (±0.53) |
|
Moryella
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.05) |
|
Mycobacterium
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.02 (±0.03) |
0.00 (±0.00) |
|
Naxibacter
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Novosphingobium
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Odoribacter
|
0.15 (±0.01) |
0.68 (±0.26) |
0.11 (±0.03) |
0.10 (±0.13) |
0.01 (±0.01) |
|
Opitutus
|
0.07 (±0.10) |
0.01 (±0.01) |
0.01 (±0.01) |
0.02 (±0.03) |
0.00 (±0.00) |
|
Paenibacillus
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.08 (±0.11) |
|
Paenisporosarcina
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.05) |
|
Paludibacter
|
0.38 (±0.17) |
0.24 (±0.02) |
0.10 (±0.05) |
0.33 (±0.14) |
0.35 (±0.10) |
|
Parabacteroides
|
0.56 (±0.11) |
1.66 (±0.51) |
0.67 (±0.39) |
2.20 (±0.21) |
3.05 (±1.27) |
|
Propionibacterium
|
0.00 (±0.00) |
0.17 (±0.21) |
0.08 (±0.12) |
0.20 (±0.06) |
0.23 (±0.08) |
|
Propionicicella
|
0.05 (±0.06) |
0.01 (±0.01) |
0.00 (±0.00) |
0.12 (±0.12) |
0.01 (±0.01) |
|
Propionivibrio
|
0.07 (±0.09) |
0.01 (±0.01) |
0.03 (±0.00) |
0.12 (±0.08) |
0.58 (±0.59) |
|
Pseudoclavibacter
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.44 (±0.62) |
|
Pseudomonas
|
0.00 (±0.00) |
0.00 (±0.00) |
0.10 (±0.14) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Rickettsiella
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.08 (±0.11) |
|
Riemerella
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.04) |
|
Roseburia
|
0.00 (±0.00) |
0.00 (±0.00) |
0.05 (±0.06) |
0.05 (±0.07) |
0.00 (±0.00) |
|
Sanguibacter
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.05) |
|
Serratia
|
10.27 (±9.45) |
15.01 (±20.00) |
0.02 (±0.03) |
0.94 (±0.03) |
1.56 (±0.15) |
|
Shimazuella
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.05 (±0.07) |
|
Shuttleworthia
|
0.00 (±0.00) |
0.05 (±0.07) |
0.01 (±0.02) |
0.27 (±0.27) |
0.02 (±0.03) |
|
Singulisphaera
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.08 (±0.11) |
0.00 (±0.00) |
|
Sphingobium
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.04) |
0.12 (±0.17) |
|
Sphingomonas
|
0.00 (±0.00) |
0.05 (±0.07) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Spirochaeta
|
0.16 (±0.23) |
0.12 (±0.02) |
0.07 (±0.08) |
0.04 (±0.04) |
0.03 (±0.02) |
|
Sporolactobacillus
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.06 (±0.06) |
0.01 (±0.02) |
|
Staphylococcus
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.04 (±0.06) |
0.00 (±0.00) |
|
Stenotrophomonas
|
0.00 (±0.00) |
0.02 (±0.03) |
0.05 (±0.07) |
0.07 (±0.00) |
0.21 (±0.27) |
|
Stenoxybacter
|
0.08 (±0.05) |
2.55 (±3.61) |
0.02 (±0.01) |
0.19 (±0.14) |
0.03 (±0.04) |
|
Streptococcus
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.05 (±0.06) |
|
Subdoligranulum
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.04) |
0.00 (±0.00) |
|
Tannerella
|
0.00 (±0.00) |
0.12 (±0.05) |
0.01 (±0.01) |
0.17 (±0.10) |
0.10 (±0.14) |
|
Telmatospirillum
|
0.00 (±0.00) |
0.01 (±0.01) |
0.00 (±0.00) |
0.00 (±0.00) |
0.08 (±0.06) |
|
Terriglobus
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Thalassospira
|
0.19 (±0.21) |
0.10 (±0.06) |
0.22 (±0.09) |
0.44 (±0.44) |
0.02 (±0.03) |
|
Thermus
|
0.00 (±0.00) |
0.00 (±0.00) |
0.03 (±0.04) |
0.30 (±0.31) |
0.00 (±0.00) |
|
Treponema
|
11.63 (±2.67) |
5.29 (±3.40) |
17.45 (±0.58) |
5.86 (±1.38) |
5.39 (±5.35) |
|
Weeksella
|
0.01 (±0.02) |
0.09 (±0.13) |
0.25 (±0.15) |
0.19 (±0.27) |
0.02 (±0.02) |
|
Weissella
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.73 (±1.03) |
0.00 (±0.00) |
|
Yersinia
|
0.06 (±0.08) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
|
Yokenella
|
0.00 (±0.00) |
0.00 (±0.00) |
0.00 (±0.00) |
0.32 (±0.10) |
0.79 (±0.54) |
|
The rest |
37.93 (±5.19) |
45.43 (±5.58) |
55.41 (±3.73) |
66.08 (±10.88) |
60.45 (±3.26) |
Download Excel Table
References
Ahn S.J., Costa J., Emanuel J.R. PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Research. 1996; 24(13):2623-2625.

Brune A. Symbiotic digestion of lignocellulose in termite guts. Nature Reviews Microbiology. 2014; 12(3):168-180.

Brune A., Ohkuma M. In: Bignell D.E., Roisin Y., Lo N., editors. Role of the termite gut microbiota in symbiotic digestion. Biology of termites: A modern synthesis. 2011; Springer. p. 439-475.

Cho M-J., Kim Y-H., Shin K., Kim Y-K., Kim Y-S., Kim T-J. Symbiotic adaptation of bacteria in the gut of
Reticulitermes speratus: Low endo-
β -1,4-glucanase activity. Biochemical and Biophysical Research Communications. 2010a; 395(3):432-435.

Cho M.J., Shin K., Kim Y-K., Kim Y-S., Kim T-J. Phylogenetic analysis of
Reticulitermes speratus using the mitochondrial cytochrome C oxidase subunit I gene. Journal of the Korean Wood Science and Technology. 2010b; 38(2):135-139.

Cleveland L.R. Symbiosis between termites and their intestinal protozoa. Proceedings of the National Academy of Sciences of the United States of America. 1923; 9(12):424-428.

Evans T.A., Forschler B.T., Grace J.K. Biology of invasive termites: A worldwide review. Annual Review of Entomology. 2013; 58(1):455-474.

Geissinger O., Herlemann D.P.R., Mörschel E., Maier U.G., Brune A. The ultramicrobacterium “
Elusimicrobium minutum” gen. nov., sp. nov., thefirst cultivated representative of the termite group 1 phylum. Applied and Environmental Microbiology. 2009; 75(9):2831-2840.

Ghaly A.E., Edwards S. Termite damage to buildings: Nature of attacks and preventive construction methods. American Journal of Engineering and Applied Sciences. 2011; 4(2):187-200.

Herlemann D.P.R., Geissinger O., Brune A. The termite group I phylum is highly diverse and widespread in the environment. Applied and Environmental Microbiology. 2007; 73(20):6682-6685.

Hongoh Y., Ohkuma M., Kudo T. Molecular analysis of bacterial microbiota in the gut of the termite
Reticulitermes speratus (Isoptera; Rhinotermitidae). FEMS Microbiology Ecology. 2003; 44(2):231-242.

Inoue T., Murashima K., Azuma J.I., Sugimoto A., Slaytor M. Cellulose and xylan utilisation in the lower termite
Reticulitermes speratus. Journal of Insect Physiology. 1997; 43(3):235-242.

Jung H-S., Choi Y., Oh J-H., Lim G-H. Recent trends in temperature and precipitation over South Korea. International Journal of Climatoogy. 2002; 22:1327-1337.

Kafri M., Metzl-Raz E., Jona G., Barkai N. The cost of protein production. Cell Reports. 2016; 14(1):22-31.

Kim M.J., Choi Y.S., Lee J., Kim J.J., Kim G.H. Molecular characteristics of subterranean termites of the genus
Reticulitermes (Isoptera: Rhinotermitidae) from Korea. Annals of the Entomological Society of America. 2012; 105(1):97-102.

Kim S.H., Chung Y.J. Ingestion toxicity of fipronil on
Reticulitermes speratus kyushuensis (Isoptera: Rhinotermitidae) and its applicability as a termite bait. Journal of the Korean Wood Science and Technology. 2017; 45(2):159-167.

Kim Y.H., Lee Y.M., Kim Y.S., Cho M.J., Shin K. Cellulase production in the digestive organs of
Reticulitermes speratus, a native termite from Milyang Korea. Journal of the Korean Wood Science and Technology. 2010; 38(5):421-428.

Mun S.P., Nicholas D.D. Effect of proanthocyanidin-rich extracts from
Pinus radiata bark on termite feeding deterrence. Journal of the Korean Wood Science and Technology. 2017; 45(6):720-727.

Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of Molecular Biology. 1970; 48(3):443-453.

Ohkuma M., Kudo T. Phylogenetic diversity of the intestinal bacterial community in the termite
Reticulitermes speratus. Applied and Environmental Microbiology. 1996; 62(2):461-468.

Park Y.C., Kitade O., Schwarz M., Kim J.P., Kim W. Intraspecific molecular phylogeny, genetic variation and phylogeography of
Reticulitermes speratus (Isoptera: Rhinotermitidae). Molecular Cell. 2006; 21(1):89-103.

Peterson B.F., Scharf M.E. Lower termite associations with microbes: Synergy, protection, and interplay. Frontiers in Microbiology. 2016; 7(422).

Peterson C., Wagner T.L., Mulrooney J.E., Shelton T.G. Subterranean termites - their prevention and control in buildings. Home and Garden Bulletin. 2006; 64:38.

Rose J.M., Caron D.A. Does low temperature constrain the growth rates of heterotrophic protists? Evidence and implications for algal blooms in cold waters. Limnology and Oceanography. 2007; 52(2):886-895.

Rosengaus R.B., Zecher C.N., Schultheis K.F., Brucker R.M., Bordenstein S.R. Disruption of the termite gut microbiota and its prolonged consequences for fitness. Applied and Environmental Microbiology. 2011; 77(13):4303-4312.

Shi Y., Huang Z., Han S., Fan S., Yang H. Phylogenetic diversity of Archaea in the intestinal tract of termites from different lineages. Journal of Basic Microbiology. 2015; 55(8):1021-1028.

Watanabe H., Tokuda G. Cellulolytic systems in insects. Annual Review of Entomology. 2010; 55(1):609-632.

Yang H., Schmitt-Wagner D., Stingl U., Brune A. Niche heterogeneity determines bacterial community structure in the termite gut (
Reticulitermes santonensis). Environmental Microbiology. 2005; 7(7):916-932.